Genomics/Epigenomics
Category: Abstract Submission
2: Genomics/Epigenomics II
17 - Single nucleotide variants in 3´ untranslated region regulatory elements are associated with altered gene expression and phenotypes
Friday, April 22, 2022
6:15 PM - 8:45 PM US MT
Poster Number: 17
Publication Number: 17.110
Publication Number: 17.110
Lindsay Romo, Boston Children's Hospital, Brookline, MA, United States; Scott Findlay, MIT, Cambridge, MA, United States; Chris Burge, Massachusetts Institute of Technology, BELMONT, MA, United States

Lindsay Romo, MD, PhD
resident
Boston Children's Hospital
Brookline, Massachusetts, United States
Presenting Author(s)
Background: Rare variants in the human genome are increasingly recognized as contributing to human disease. One class of poorly understood variants is in 3´ untranslated regions (3´UTRs). Much of the regulation in 3’ UTRs occurs via interactions at RNA binding protein (RBP) and microRNA (miRNA) sites, affecting the processing of transcripts. We have found negative selection against 3´UTR variants in high-confidence RBP sites and conserved polyA signals is of similar magnitude to selection against nonsynonymous coding region variants. Examination of expression quantitative trait loci (eQTL) and genome wide association studies (GWAS) can be used to predict the functional impact of these variants.
Objective: To determine whether 3´UTR variants in regulatory elements are likely to impact gene expression and phenotype.
Design/Methods: A variant's posterior inclusion probability (PIP) is the probability that it alters gene expression (eQTL) or phenotype (GWAS). We compared the PIPs of Genotype-Tissue Expression (GTEx) project eQTLs and UK Biobank GWAS variants in 3´UTR regulatory elements to variants not in these elements. RBP sites were identified computationally and via enhanced crosslinking and immunoprecipitation (eCLIP). We contrasted allele-specific RBP binding at variants in RBP motifs to those not in motifs using the Binding Estimation of Allele-specific Protein–RNA interaction (BEAPR) method.
Results: eQTL and GWAS variants are more likely than non-eQTL and non-GWAS variants to occur in RBP motifs, eCLIP peaks, and microRNA sites, and this enrichment increases with increasing PIP. Conversely, variants in RBP motifs, eCLIP peaks, and microRNA motifs have higher mean PIPs as well as a higher proportion of high-confidence (PIP>0.5) causal variants than control variants. PIP increases with increasing numbers of RBP motifs or eCLIP peaks, or as the predicted strength of the microRNA binding site increases. Variant PIP is also related to the number of alternatively polyadenylated isoforms. Variants in common 3´UTR regions of genes with fewer isoforms have higher PIPs, especially if proximal ( < 50 nucleotides) to the polyA site. Variants in RBP motifs are more than four times more likely to exhibit allele-specific RBP binding than those not in motifs.Conclusion(s): These results yield a model in which eQTL and GWAS variant PIP is negatively correlated with isoform number and positively correlated with microRNA motifs, predicted RBP motifs, eCLIP peaks, and proximity to poly A sites, suggesting rare 3´UTR variants in these regions are under negative selection due to their impact on gene expression and phenotype.
Methods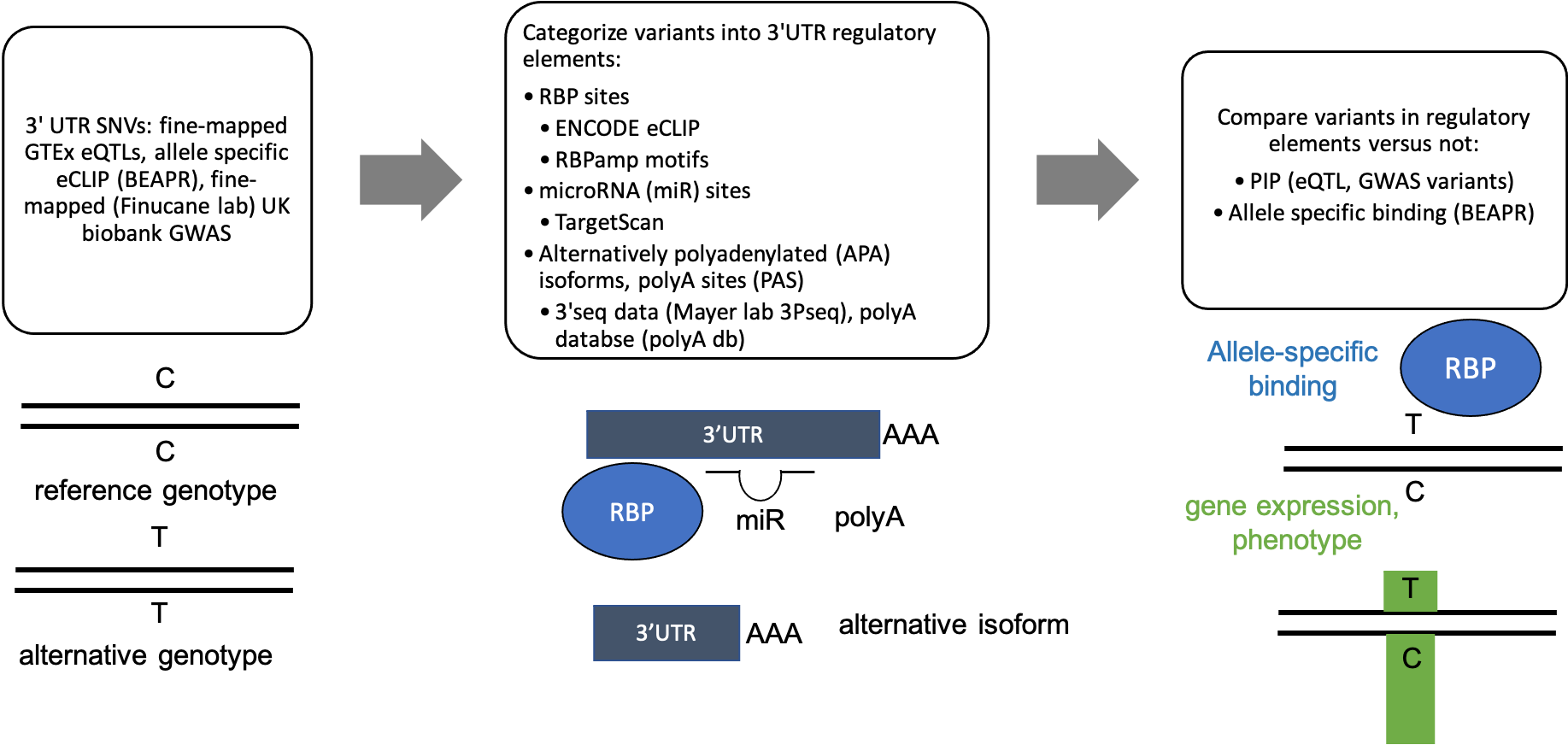 3'UTR variant analysis pipeline to assess impact of variants on RBP binding, gene expression, phenotype.
3'UTR variant analysis pipeline to assess impact of variants on RBP binding, gene expression, phenotype.
Objective: To determine whether 3´UTR variants in regulatory elements are likely to impact gene expression and phenotype.
Design/Methods: A variant's posterior inclusion probability (PIP) is the probability that it alters gene expression (eQTL) or phenotype (GWAS). We compared the PIPs of Genotype-Tissue Expression (GTEx) project eQTLs and UK Biobank GWAS variants in 3´UTR regulatory elements to variants not in these elements. RBP sites were identified computationally and via enhanced crosslinking and immunoprecipitation (eCLIP). We contrasted allele-specific RBP binding at variants in RBP motifs to those not in motifs using the Binding Estimation of Allele-specific Protein–RNA interaction (BEAPR) method.
Results: eQTL and GWAS variants are more likely than non-eQTL and non-GWAS variants to occur in RBP motifs, eCLIP peaks, and microRNA sites, and this enrichment increases with increasing PIP. Conversely, variants in RBP motifs, eCLIP peaks, and microRNA motifs have higher mean PIPs as well as a higher proportion of high-confidence (PIP>0.5) causal variants than control variants. PIP increases with increasing numbers of RBP motifs or eCLIP peaks, or as the predicted strength of the microRNA binding site increases. Variant PIP is also related to the number of alternatively polyadenylated isoforms. Variants in common 3´UTR regions of genes with fewer isoforms have higher PIPs, especially if proximal ( < 50 nucleotides) to the polyA site. Variants in RBP motifs are more than four times more likely to exhibit allele-specific RBP binding than those not in motifs.Conclusion(s): These results yield a model in which eQTL and GWAS variant PIP is negatively correlated with isoform number and positively correlated with microRNA motifs, predicted RBP motifs, eCLIP peaks, and proximity to poly A sites, suggesting rare 3´UTR variants in these regions are under negative selection due to their impact on gene expression and phenotype.
Methods
 3'UTR variant analysis pipeline to assess impact of variants on RBP binding, gene expression, phenotype.
3'UTR variant analysis pipeline to assess impact of variants on RBP binding, gene expression, phenotype.
Designed by Cadmium
|Technical Support
© Copyright 2025 Cadmium. All Rights Reserved.
