Neonatal Neurology: Basic
Category: Abstract Submission
Neurology 3: Basic-Translational
417 - Next Generation RNA Sequencing Provides Novel Insights into the Mechanisms of Brain injury in Neonatal Hypoxic-Ischemic Encephalopathy
Saturday, April 23, 2022
3:30 PM - 6:00 PM US MT
Poster Number: 417
Publication Number: 417.237
Publication Number: 417.237
Angela Saadat, Eastern Virginia Medical School, Norfolk, VA, United States; Haree Pallera, Eastern Virginia Medical School, Virginia Beach, VA, United States; Heong Jin C. Ahn, ACOM, Dothan, AL, United States; Tushar A. Shah, Eastern Virginia Medical School, Norfolk, VA, United States

Tushar A. Shah, MD MPH
Associate Professor of Pediatrics
Eastern Virginia Medical School
Norfolk, Virginia, United States
Presenting Author(s)
Background: Therapeutic hypothermia (TH) is the only proven treatment known to improve outcomes in neonatal hypoxic-ischemic encephalopathy (HIE). Due to clinical heterogeneity, hypothermic neuroprotection is not universal, with 50% of cooled babies developing adverse neurodevelopmental outcomes. Several therapeutic agents that have shown promise in the treatment of HIE have failed to synergize with TH. Transcriptomic profiling and differential gene expression (DGE) using Next-generation RNA-sequencing (NGS) could identify key pathways instrumental in neonatal HIE, and the modulatory effect of TH on these pathways
Objective: To interrogate the heterogeneity of the therapeutic response to TH in neonatal HIE using NGS, with the goal of identifying key pathways that can be targeted for therapy
Design/Methods: The Vannucci model was used to induce HIE in term-equivalent rat pups (P10-12), and randomized into Sham (control), Normothermia (NT) and Hypothermia (TH) groups (n=4 per group per time point). Animals were harvested 24h and 5d after injury. Purified whole-brain RNA was sequenced using Illumina short-read HiSeq, 2x150bp configuration. Using DESeq2, a comparison of gene expression between the groups was performed. Genes with an adjusted p-value < 0.05 and absolute log2 fold change > 1 (Wald test) were called as DEGs for each comparison. Gprofiler, GSea, Reactome and Cytoscape were used to analyze gene ontology (GO) and identify significantly enriched pathways.
Results: Fig. 1 (A, B) shows number of significantly up and downregulated genes between groups. 296 genes were significantly upregulated and 402 genes were significantly downregulated in the HT group 24h after injury, when compared to the NT group. Fig.1 (C-H) shows comparative heatmaps of top up and downregulated genes. Significant DEGs were clustered by their GO. TH animals demonstrated maximal enrichment in pathways affecting calcium signaling (upregulation) and Cell cycle and complement cascades (downregulation) 24h after injury, compared to NT (Table 1). Compared to controls, NT animals demonstrated maximal enrichment in pathways affecting innate immunity (upregulation) and cholesterol biosynthesis (downregulation) 24h after injury (Fig.3). 5 days after injury, NT animals demonstrate a recovery in cholesterol biosynthesisConclusion(s): NGS demonstrates the complexity, heterogeneity and variability of cellular responses to HIE and TH. Further elucidating the importance of key pathways of cholesterol biosynthesis and innate immune response may aid in developing an effective therapeutic adjunct to TH
Differential gene expression between groups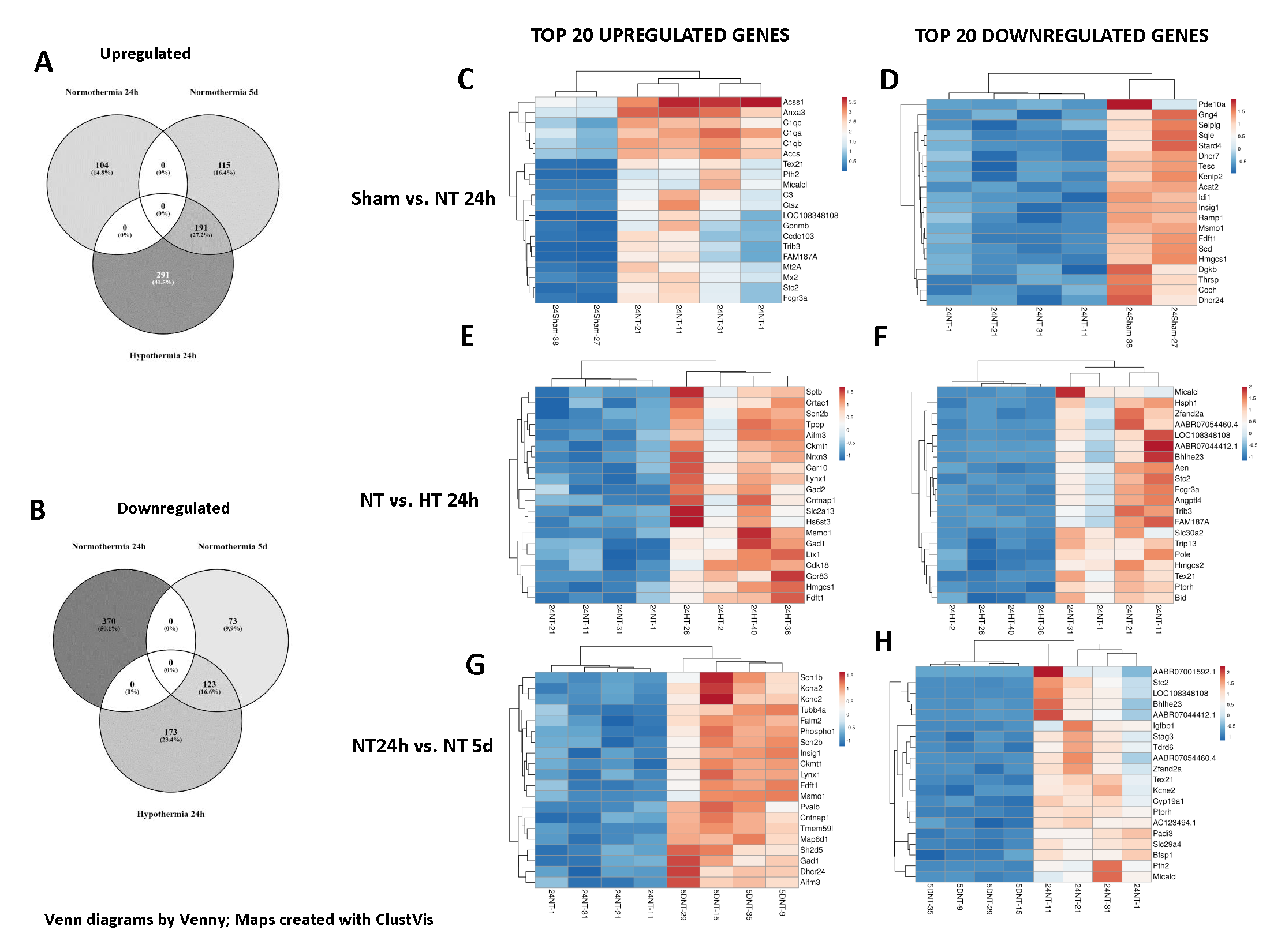 Total number of upregulated (A) and downregulated (B) genes between groups is demonstrated. There is significant overlap between hypothermia animals 24h after injury and NT animals 5d after injury (also seen in F and H), possibly indicating that HT accelerated certain reparative processes. Heatmaps demonstrate the top upregulated and downregulated genes in NT animals 24 after injury when compared to Sham (C,D), HT animals 24h after injury when compared to NT 24h after injury (E,F) and NT animals 5d after injury when compared to NT animals 24h after injury (G,H). Overall, gene enrichment was seen in pathways affecting Steroid/Cholesterol biosynthesis, Complement activation, Neutrophil/astrocyte activation and Neurotransmitter (GABA) biosynthesis. (Venn diagrams generated on Venny, Heatmaps generated on ClustVis)
Total number of upregulated (A) and downregulated (B) genes between groups is demonstrated. There is significant overlap between hypothermia animals 24h after injury and NT animals 5d after injury (also seen in F and H), possibly indicating that HT accelerated certain reparative processes. Heatmaps demonstrate the top upregulated and downregulated genes in NT animals 24 after injury when compared to Sham (C,D), HT animals 24h after injury when compared to NT 24h after injury (E,F) and NT animals 5d after injury when compared to NT animals 24h after injury (G,H). Overall, gene enrichment was seen in pathways affecting Steroid/Cholesterol biosynthesis, Complement activation, Neutrophil/astrocyte activation and Neurotransmitter (GABA) biosynthesis. (Venn diagrams generated on Venny, Heatmaps generated on ClustVis)
Clustering significant differentially expressed genes by their Gene Ontology and Key Pathways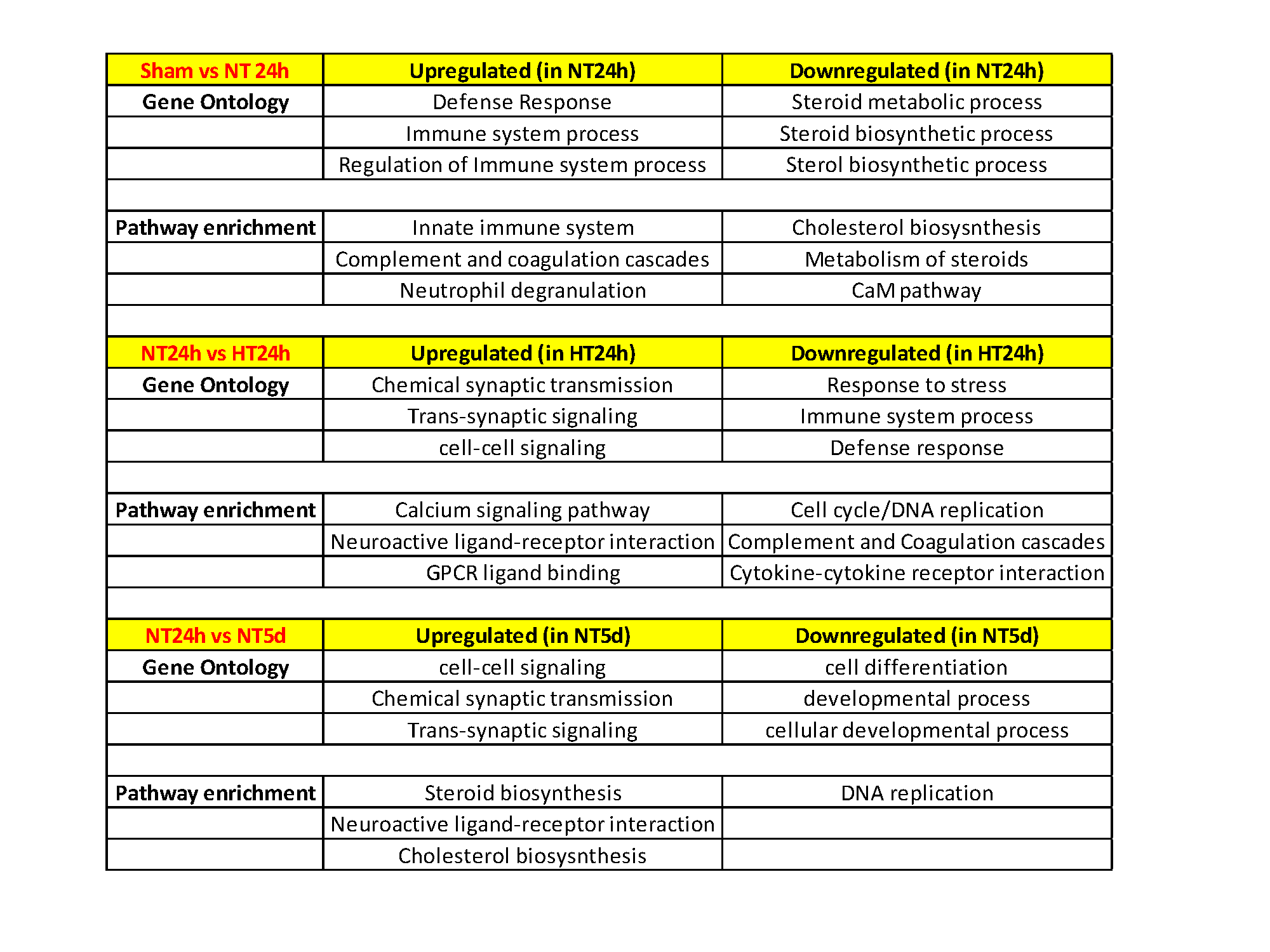 Detailed gene expression comparison between groups identifies key pathways that may be potential therapeutic targets or biomarkers for disease progression/prognosis - Innate immunity, Steroid/cholesterol biosynthesis and Chemical synaptic transmission pathways.
Detailed gene expression comparison between groups identifies key pathways that may be potential therapeutic targets or biomarkers for disease progression/prognosis - Innate immunity, Steroid/cholesterol biosynthesis and Chemical synaptic transmission pathways.
Objective: To interrogate the heterogeneity of the therapeutic response to TH in neonatal HIE using NGS, with the goal of identifying key pathways that can be targeted for therapy
Design/Methods: The Vannucci model was used to induce HIE in term-equivalent rat pups (P10-12), and randomized into Sham (control), Normothermia (NT) and Hypothermia (TH) groups (n=4 per group per time point). Animals were harvested 24h and 5d after injury. Purified whole-brain RNA was sequenced using Illumina short-read HiSeq, 2x150bp configuration. Using DESeq2, a comparison of gene expression between the groups was performed. Genes with an adjusted p-value < 0.05 and absolute log2 fold change > 1 (Wald test) were called as DEGs for each comparison. Gprofiler, GSea, Reactome and Cytoscape were used to analyze gene ontology (GO) and identify significantly enriched pathways.
Results: Fig. 1 (A, B) shows number of significantly up and downregulated genes between groups. 296 genes were significantly upregulated and 402 genes were significantly downregulated in the HT group 24h after injury, when compared to the NT group. Fig.1 (C-H) shows comparative heatmaps of top up and downregulated genes. Significant DEGs were clustered by their GO. TH animals demonstrated maximal enrichment in pathways affecting calcium signaling (upregulation) and Cell cycle and complement cascades (downregulation) 24h after injury, compared to NT (Table 1). Compared to controls, NT animals demonstrated maximal enrichment in pathways affecting innate immunity (upregulation) and cholesterol biosynthesis (downregulation) 24h after injury (Fig.3). 5 days after injury, NT animals demonstrate a recovery in cholesterol biosynthesisConclusion(s): NGS demonstrates the complexity, heterogeneity and variability of cellular responses to HIE and TH. Further elucidating the importance of key pathways of cholesterol biosynthesis and innate immune response may aid in developing an effective therapeutic adjunct to TH
Differential gene expression between groups
 Total number of upregulated (A) and downregulated (B) genes between groups is demonstrated. There is significant overlap between hypothermia animals 24h after injury and NT animals 5d after injury (also seen in F and H), possibly indicating that HT accelerated certain reparative processes. Heatmaps demonstrate the top upregulated and downregulated genes in NT animals 24 after injury when compared to Sham (C,D), HT animals 24h after injury when compared to NT 24h after injury (E,F) and NT animals 5d after injury when compared to NT animals 24h after injury (G,H). Overall, gene enrichment was seen in pathways affecting Steroid/Cholesterol biosynthesis, Complement activation, Neutrophil/astrocyte activation and Neurotransmitter (GABA) biosynthesis. (Venn diagrams generated on Venny, Heatmaps generated on ClustVis)
Total number of upregulated (A) and downregulated (B) genes between groups is demonstrated. There is significant overlap between hypothermia animals 24h after injury and NT animals 5d after injury (also seen in F and H), possibly indicating that HT accelerated certain reparative processes. Heatmaps demonstrate the top upregulated and downregulated genes in NT animals 24 after injury when compared to Sham (C,D), HT animals 24h after injury when compared to NT 24h after injury (E,F) and NT animals 5d after injury when compared to NT animals 24h after injury (G,H). Overall, gene enrichment was seen in pathways affecting Steroid/Cholesterol biosynthesis, Complement activation, Neutrophil/astrocyte activation and Neurotransmitter (GABA) biosynthesis. (Venn diagrams generated on Venny, Heatmaps generated on ClustVis)Clustering significant differentially expressed genes by their Gene Ontology and Key Pathways
 Detailed gene expression comparison between groups identifies key pathways that may be potential therapeutic targets or biomarkers for disease progression/prognosis - Innate immunity, Steroid/cholesterol biosynthesis and Chemical synaptic transmission pathways.
Detailed gene expression comparison between groups identifies key pathways that may be potential therapeutic targets or biomarkers for disease progression/prognosis - Innate immunity, Steroid/cholesterol biosynthesis and Chemical synaptic transmission pathways.