Back
Genomics/Epigenomics
Category: Abstract Submission
2: Genomics/Epigenomics II
10 - A Functional Genomics Approach to Identifying Candidate Target Genes on JIA Risk Haplotypes
Friday, April 22, 2022
6:15 PM – 8:45 PM US MT
Poster Number: 10
Publication Number: 10.110
Publication Number: 10.110
Emma K. Haley, Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo, Buffalo, NY, United States; Kaiyu Jiang, Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo, Buffalo, NY, United States; James N. Jarvis, Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo, Buffalo, NY, United States

Jim N. Jarvis, MD
Professor of Pediatrics
Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo
Buffalo, New York, United States
Presenting Author(s)
Background: GWAS have identified multiple regions that contribute to genetic risk for juvenile idiopathic arthritis, but neither the causal variants nor their target genes are known. We have shown that JIA risk loci are enriched for epigenetic marks associated with enhancer function (H3K4me1 and/or H3K27ac) in neutrophils and CD4+ T cells. However, H3K4me1/H3K27ac marks are not prima facie evidence that a given region has enhancer function.
Objective: To identify relevant enhancers and target genes on JIA risk haplotypes.
Design/Methods: We queried 23 JIA risk haplotypes. Using BedTools, we queried Global Run-On-Sequencing (GRO-seq) data, to identify bidirectional RNA synthesis typical of functional enhancers, in CD4+ T cells. We examined these same regions for the presence of H3K27ac peaks and transcription-factor binding sites using ChIP-Seq. For these candidate enhancers, genes in their respective topologically associated domains were identified using publicly available capture HiC. Next, we used CRISPRi to interrogate TYK2 and IRF1 enhancer candidates to identify the target genes of these enhancers. We used Jurkat cells with the dCas9-KRAB epigenome editing construct stably integrated into the genome. Similarly, sgRNA plasmids were stably transfected into cells using the hypBase system, and dCas-KRAB induced using doxycline. Candidate target gene expression was determined by qPCR.
Results: We identified GROseq peaks on 15 of the 23 JIA risk haplotypes queried. These peaks occurred at a great frequency that would be expected by chance (p=0.0002). The 15 risk haplotypes contained 39 GRO-seq peaks; 24 of these GRO-seq peaks were also flanked by H3K27ac ChIP-seq peaks. Taking into account transcription-factor binding and DNAse hypersensitivity, the list of potential enhancers on the JIA risk haplotypes was narrowed down to 17 candidates. We chose TYK2 and IRF1 regions for additional functional in vitro analysis based on their relevance to autoimmune diseases and the expression levels of candidate target genes in Jurkat cells. After ablating the TYK2 enhancer with dCas-KRAB, we found reduced expression of CDC37, but not TYK2, ILF3, or ICAM4, which are in the same chromatin loop. Attenuating the IRF1 enhancer reduced expression of IRF1, KIF3A, and C5orf56, but not GDF9.Conclusion(s): We demonstrate a systematic approach for identifying target genes on JIA genetic risk haplotypes. Using functional data sets and knowledge of 3D chromatin structures, we identified a finite set of target genes in the IRF1 and TYK2 risk loci, some of which are not situated on the disease haplotypes.
Identification of genes regulated by an intergenic enhancer in the IRF1 locus using CRISPRi.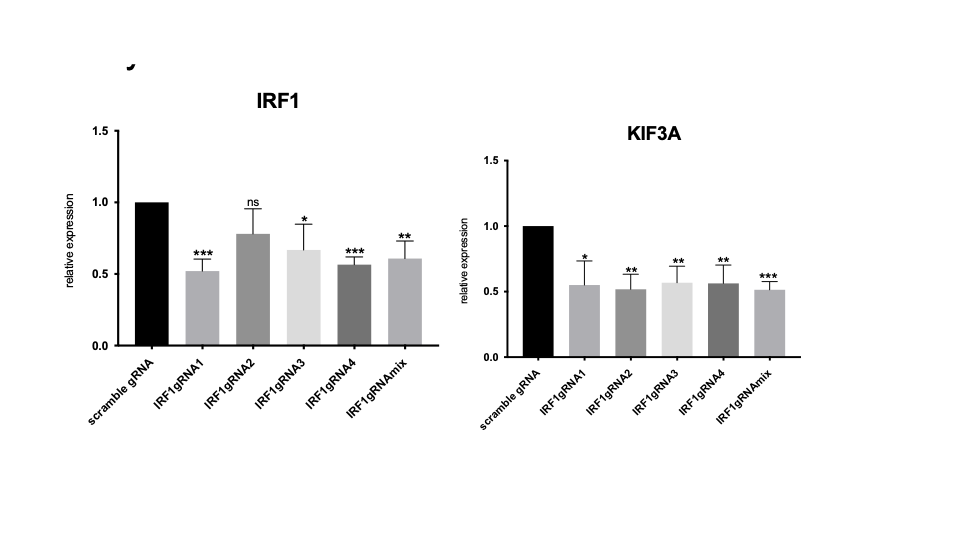 The bar graph showing results obtained by ablating the intergenic enhancer harboring rs2706385 and rs2706386 on the IRF1 locus using CRISPRi with dCas-KRAB and 4 gRNAs tiled across the enhancer region or used together (“gRNAmix”). Ablating this enhancer reduces expression of both IRF1and KIF3A. *p < 0.05; **p < 0.01; ***p < 0.001.Ns=Not significant. Attenuating this enhancer had no effect on the expression of GDF9 (not shown), another gene within the same TAD as this enhancer.
The bar graph showing results obtained by ablating the intergenic enhancer harboring rs2706385 and rs2706386 on the IRF1 locus using CRISPRi with dCas-KRAB and 4 gRNAs tiled across the enhancer region or used together (“gRNAmix”). Ablating this enhancer reduces expression of both IRF1and KIF3A. *p < 0.05; **p < 0.01; ***p < 0.001.Ns=Not significant. Attenuating this enhancer had no effect on the expression of GDF9 (not shown), another gene within the same TAD as this enhancer.
Objective: To identify relevant enhancers and target genes on JIA risk haplotypes.
Design/Methods: We queried 23 JIA risk haplotypes. Using BedTools, we queried Global Run-On-Sequencing (GRO-seq) data, to identify bidirectional RNA synthesis typical of functional enhancers, in CD4+ T cells. We examined these same regions for the presence of H3K27ac peaks and transcription-factor binding sites using ChIP-Seq. For these candidate enhancers, genes in their respective topologically associated domains were identified using publicly available capture HiC. Next, we used CRISPRi to interrogate TYK2 and IRF1 enhancer candidates to identify the target genes of these enhancers. We used Jurkat cells with the dCas9-KRAB epigenome editing construct stably integrated into the genome. Similarly, sgRNA plasmids were stably transfected into cells using the hypBase system, and dCas-KRAB induced using doxycline. Candidate target gene expression was determined by qPCR.
Results: We identified GROseq peaks on 15 of the 23 JIA risk haplotypes queried. These peaks occurred at a great frequency that would be expected by chance (p=0.0002). The 15 risk haplotypes contained 39 GRO-seq peaks; 24 of these GRO-seq peaks were also flanked by H3K27ac ChIP-seq peaks. Taking into account transcription-factor binding and DNAse hypersensitivity, the list of potential enhancers on the JIA risk haplotypes was narrowed down to 17 candidates. We chose TYK2 and IRF1 regions for additional functional in vitro analysis based on their relevance to autoimmune diseases and the expression levels of candidate target genes in Jurkat cells. After ablating the TYK2 enhancer with dCas-KRAB, we found reduced expression of CDC37, but not TYK2, ILF3, or ICAM4, which are in the same chromatin loop. Attenuating the IRF1 enhancer reduced expression of IRF1, KIF3A, and C5orf56, but not GDF9.Conclusion(s): We demonstrate a systematic approach for identifying target genes on JIA genetic risk haplotypes. Using functional data sets and knowledge of 3D chromatin structures, we identified a finite set of target genes in the IRF1 and TYK2 risk loci, some of which are not situated on the disease haplotypes.
Identification of genes regulated by an intergenic enhancer in the IRF1 locus using CRISPRi.
 The bar graph showing results obtained by ablating the intergenic enhancer harboring rs2706385 and rs2706386 on the IRF1 locus using CRISPRi with dCas-KRAB and 4 gRNAs tiled across the enhancer region or used together (“gRNAmix”). Ablating this enhancer reduces expression of both IRF1and KIF3A. *p < 0.05; **p < 0.01; ***p < 0.001.Ns=Not significant. Attenuating this enhancer had no effect on the expression of GDF9 (not shown), another gene within the same TAD as this enhancer.
The bar graph showing results obtained by ablating the intergenic enhancer harboring rs2706385 and rs2706386 on the IRF1 locus using CRISPRi with dCas-KRAB and 4 gRNAs tiled across the enhancer region or used together (“gRNAmix”). Ablating this enhancer reduces expression of both IRF1and KIF3A. *p < 0.05; **p < 0.01; ***p < 0.001.Ns=Not significant. Attenuating this enhancer had no effect on the expression of GDF9 (not shown), another gene within the same TAD as this enhancer.