Neonatal GI Physiology & NEC
Category: Abstract Submission
Neonatal GI Physiology & NEC III
513 - Spatial single cell analysis reveals altered signaling and interactions in regulatory T lymphocytes in NEC
Monday, April 25, 2022
3:30 PM - 6:00 PM US MT
Poster Number: 513
Bunmi Olaloye, Yale School of Medicine, NEW HAVEN, CT, United States; Adi Egozi, Weizmann Institute of Science, Rehovot, Tel Aviv, Israel; Tatiana Silva, Yale School of Medicine, New Haven, CT, United States; Shalev Itzkovitz, Weizmann Institute of Science, rehovot, HaDarom, Israel; Liza Konnikova, Yale University School of Medicine, New Haven, CT, United States
.jpg)
Bunmi Olaloye, M.D
Neonatology Attending/Post Doctoral Fellow
Yale School of Medicine
NEW HAVEN, Connecticut, United States
Presenting Author(s)
Background: Infants born extremely prematurely are susceptible to necrotizing enterocolitis (NEC). This devastating disease remains poorly understood and treatment is non-specific. There is an urgent need to identify biomarkers and to develop targeted therapies
Objective: Single cell analysis with a spatial resolution of NEC-affected mucosa can identify altered signaling and cellular interactions that could serve as potential therapeutic targets.
Design/Methods: Small intestinal tissue samples obtained at initial surgery for NEC (n=20, gestational age (GA) 23-39 weeks) and in neonates with non-immune congenital anomalies (neonatal n=13, GA 34-40 weeks) were compared. Single-cell RNA sequencing (scRNAseq), deconvolution of bulk sequencing data, and imaging mass cytometry (IMC) were performed (Fig 1a).
Results: We identified 7 distinct populations of lymphocytes by scRNAseq (Fig 1b-c). Analysis of differentially expressed genes showed that genes required for appropriate T reg suppressive function (FOXP3, CTLA4) were decreased and SELL (CD62L), CCR7, SOCS1 and a number of ribosomal genes were increased in NEC compard to neonates (Fig 1d). This is consistent with upregulation of SOCS1, a downstream target of STAT3 signaling and IMC analysis showed an increase in pSTAT3 in NEC associated Tregs (Fig 2a-c). Interactions between T regs and macrophages were altered. T regs in NEC signaled to macrophages through IL1R1/IL1B, CAV1-TGFB1 and ABCA1-CALM1 (Fig 1f). Furthermore, IMC highlighted altered interactions in NEC Tregs. Specifically, decreased interaction with other T cells (p < 0.01), and increased interaction with inflammatory macrophages (Fig 2f, p< 0.01). Conclusion(s): In summary, we characterize altered cellular interactions and transcriptional signatures in NEC-associated regulatory T cells that contribute to disease pathogenesis and could serve as potential targets for future therapies.
In summary, we characterize altered cellular interactions and transcriptional signatures in NEC-associated regulatory T cells that contribute to disease pathogenesis and could serve as potential targets for future therapies.
Figure 1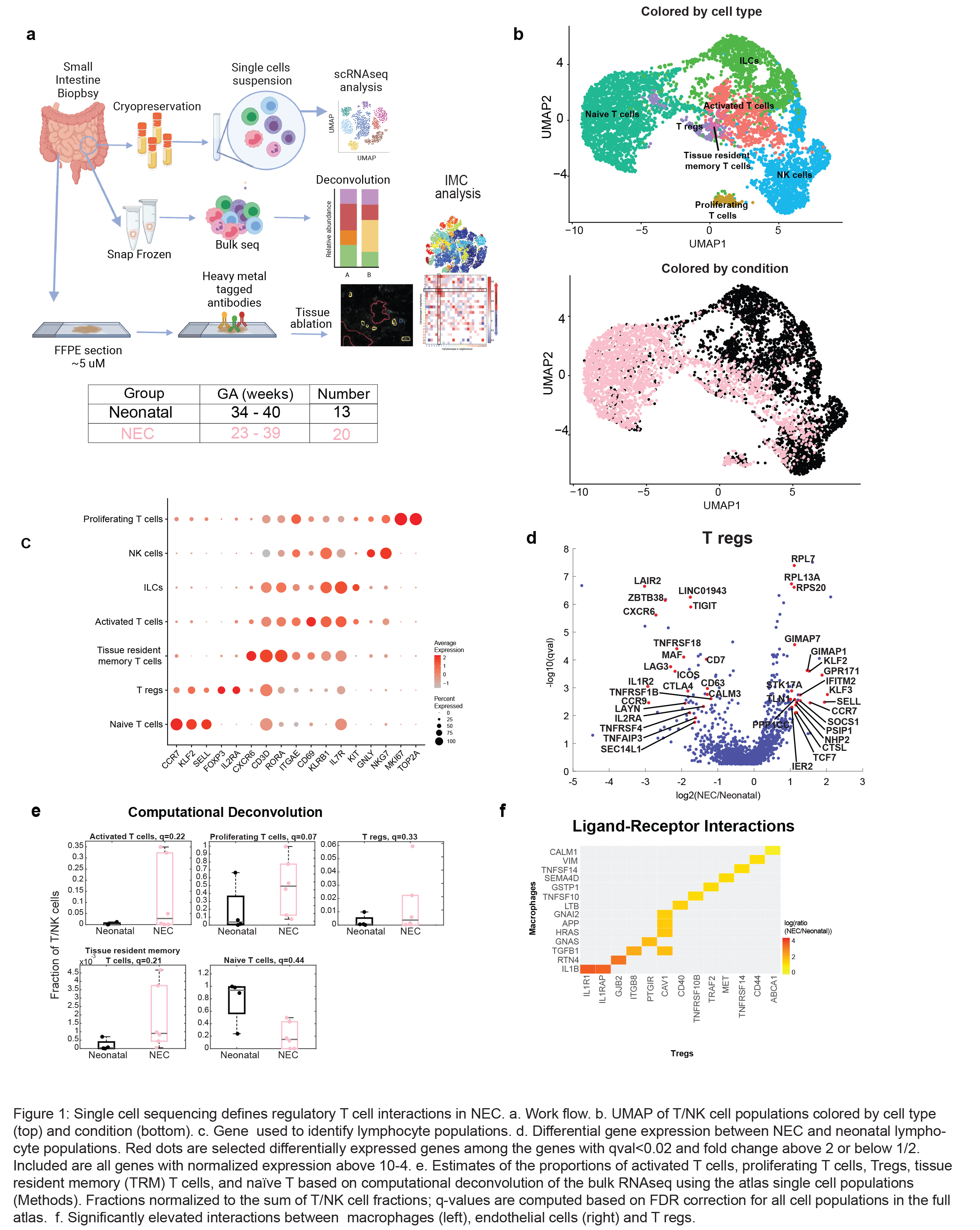
Figure 2.jpg) Figure 2: Imaging mass cytometry identifies altered signaling and cellular intereactions in Tregs in NEC. a-b. t-stochastic neighborhood embedding of all cells identified in NEC (n=5) and neonatal (n=3) samples with Phenograph analysis on Histocat (Schapiro et al 2017) displayed by cell (a) and by group (b). c. T cells are reduced in number and with increased expression of pSTAT3. d. How row and column interactions are defined in e. e. Cellular intereactions are reduced in NEC T regs compared to Neonatal. Gray stars represent intereactions in NEC that are absent in neonates. Black starts (*) represent interactions in neonatal samples but not in NEC. f. Representative images of Treg intereactions in NEC. Each dot represents 1 image, 2 images analyzed/sample. Mann-Whitney U test. *p-value 0.05.
Figure 2: Imaging mass cytometry identifies altered signaling and cellular intereactions in Tregs in NEC. a-b. t-stochastic neighborhood embedding of all cells identified in NEC (n=5) and neonatal (n=3) samples with Phenograph analysis on Histocat (Schapiro et al 2017) displayed by cell (a) and by group (b). c. T cells are reduced in number and with increased expression of pSTAT3. d. How row and column interactions are defined in e. e. Cellular intereactions are reduced in NEC T regs compared to Neonatal. Gray stars represent intereactions in NEC that are absent in neonates. Black starts (*) represent interactions in neonatal samples but not in NEC. f. Representative images of Treg intereactions in NEC. Each dot represents 1 image, 2 images analyzed/sample. Mann-Whitney U test. *p-value 0.05.
Objective: Single cell analysis with a spatial resolution of NEC-affected mucosa can identify altered signaling and cellular interactions that could serve as potential therapeutic targets.
Design/Methods: Small intestinal tissue samples obtained at initial surgery for NEC (n=20, gestational age (GA) 23-39 weeks) and in neonates with non-immune congenital anomalies (neonatal n=13, GA 34-40 weeks) were compared. Single-cell RNA sequencing (scRNAseq), deconvolution of bulk sequencing data, and imaging mass cytometry (IMC) were performed (Fig 1a).
Results: We identified 7 distinct populations of lymphocytes by scRNAseq (Fig 1b-c). Analysis of differentially expressed genes showed that genes required for appropriate T reg suppressive function (FOXP3, CTLA4) were decreased and SELL (CD62L), CCR7, SOCS1 and a number of ribosomal genes were increased in NEC compard to neonates (Fig 1d). This is consistent with upregulation of SOCS1, a downstream target of STAT3 signaling and IMC analysis showed an increase in pSTAT3 in NEC associated Tregs (Fig 2a-c). Interactions between T regs and macrophages were altered. T regs in NEC signaled to macrophages through IL1R1/IL1B, CAV1-TGFB1 and ABCA1-CALM1 (Fig 1f). Furthermore, IMC highlighted altered interactions in NEC Tregs. Specifically, decreased interaction with other T cells (p < 0.01), and increased interaction with inflammatory macrophages (Fig 2f, p< 0.01). Conclusion(s): In summary, we characterize altered cellular interactions and transcriptional signatures in NEC-associated regulatory T cells that contribute to disease pathogenesis and could serve as potential targets for future therapies.
In summary, we characterize altered cellular interactions and transcriptional signatures in NEC-associated regulatory T cells that contribute to disease pathogenesis and could serve as potential targets for future therapies.
Figure 1

Figure 2
.jpg) Figure 2: Imaging mass cytometry identifies altered signaling and cellular intereactions in Tregs in NEC. a-b. t-stochastic neighborhood embedding of all cells identified in NEC (n=5) and neonatal (n=3) samples with Phenograph analysis on Histocat (Schapiro et al 2017) displayed by cell (a) and by group (b). c. T cells are reduced in number and with increased expression of pSTAT3. d. How row and column interactions are defined in e. e. Cellular intereactions are reduced in NEC T regs compared to Neonatal. Gray stars represent intereactions in NEC that are absent in neonates. Black starts (*) represent interactions in neonatal samples but not in NEC. f. Representative images of Treg intereactions in NEC. Each dot represents 1 image, 2 images analyzed/sample. Mann-Whitney U test. *p-value 0.05.
Figure 2: Imaging mass cytometry identifies altered signaling and cellular intereactions in Tregs in NEC. a-b. t-stochastic neighborhood embedding of all cells identified in NEC (n=5) and neonatal (n=3) samples with Phenograph analysis on Histocat (Schapiro et al 2017) displayed by cell (a) and by group (b). c. T cells are reduced in number and with increased expression of pSTAT3. d. How row and column interactions are defined in e. e. Cellular intereactions are reduced in NEC T regs compared to Neonatal. Gray stars represent intereactions in NEC that are absent in neonates. Black starts (*) represent interactions in neonatal samples but not in NEC. f. Representative images of Treg intereactions in NEC. Each dot represents 1 image, 2 images analyzed/sample. Mann-Whitney U test. *p-value 0.05.