Genomics/Epigenomics
Category: Abstract Submission
2: Genomics/Epigenomics II
12 - A Systematic Approach for Identifying Causal Variants and Their Target Genes on JIA Risk Haplotypes
Friday, April 22, 2022
6:15 PM - 8:45 PM US MT
Poster Number: 12
Publication Number: 12.110
Publication Number: 12.110
Kaiyu Jiang, Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo, Buffalo, NY, United States; James N. Jarvis, Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo, Buffalo, NY, United States

Jim N. Jarvis, MD
Professor of Pediatrics
Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo
Buffalo, New York, United States
Presenting Author(s)
Background: GWAS have identified multiple regions of genetic risk for juvenile idiopathic arthritis (JIA). However, identifying the SNPs that drive disease risk is complicated by the fact that the SNPs used to identify risk loci are in LD with hundreds of other SNPs. Since the causal SNPs remain unknown, it is difficult to identify target genes and use genetic information to elucidate disease biology and inform patient care.
Objective: To develop a functional genomics pipeline to identify causal variants and their target genes on JIA risk haplotypes.
Design/Methods: We designed 180 bp oligonucleotides (“oligos”) representing 7,312 SNPs in LD with tag SNPs on JIA risk loci. Oligos were bar coded and cloned into a GFP-carrying plasmid vector behind a minimal promoter. We transfected oligo libraries into myeloid K562 cells to conduct a massively parallel reporter assay (MPRA) in which GFP expression for each SNP was compared to the common allele. We used 3D chromatin data to identify the chromatin loops harboring the enhancers carrying SNPs screened on MPRA. We used K562 cells stably transfected with the epigenome editing enzyme dCas-KRAB and gRNAs across the enhancer regions in the TRAF1 and LNPEP/ERAP2 loci, inducing dCas-KRAB with doxycycline and using qrtPCR to query expression levels of genes within the same chromatin loops.
Results: We identified n=44 SNPs that showed a significant difference in gene expression (FC > 2.0, FDR = 0.05) compared to the common allele. After stimulating K562 cells with IFNγ (250 ng/ml), we identified an additional 42 SNPs not identified in unstimulated cells. In many cases, we identified multiple alleles on the same haplotype, although these alleles were not always within the same functional element (e.g., intergenic vs intronic enhancers). Ablating the intergenic enhancer harboring rs10985080 on the TRAF1 haplotype using CRISPRi significantly reduced the expression of PHF1, but not C5, which is in the same chromatin loop. Using a similar approach for an intergenic enhancer in the LNPEP/ERAP2 locus, we found reduced expression of both LNPEP and ERAP2 but not CAST.Conclusion(s): We demonstrate a systematic method for identifying causal variants on JIA risk haplotypes based on observed functional properties. We also demonstrate that, once these variants are identified, target genes can be quickly identified using both publicly available 3D chromatin data and additional functional assays such as CRISPRi.
CRISPRi identifies target genes of enhancers harboring JIA-associated variants detected on MPRA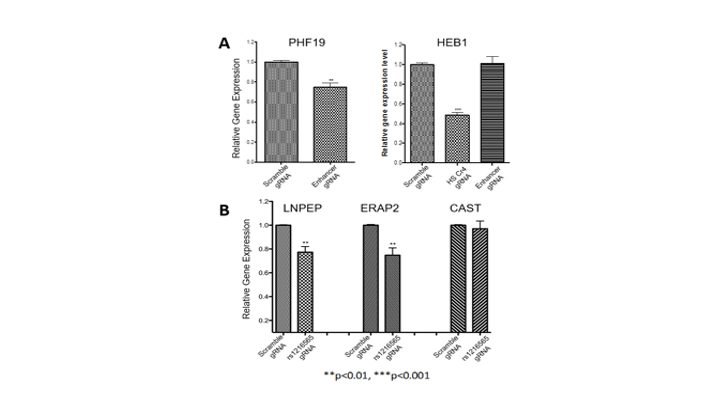 For each experiment, 4 gRNAs were targeted to the functional regions of intergenic enhancers in the TRAF1 locus and the ERAP2/LNPEP locus. Bar graphs summarize the results of 4 independent experiments. (A) Attenuation of the intergenic enhancer at the TRAF1 locus significantly reduced expression of PHF1, but not C5 (not shown). Off-target effects are monitored by showing that these gRNAs have no effect on HEB1 expression, but that gRNAs directed to an enhancer known to regulated HEB1 attenuates expression. (B) Attenuating the intergenic enhancer in the LNPEP/ERAP1 locus reduces expression of both LNPEP and ERAP2, but not the adjacent gene, CAST, nor the HEB1 gene (not shown). In each experiment, scrambled versions of the gRNAs had no effect on expression of the putative targets.
For each experiment, 4 gRNAs were targeted to the functional regions of intergenic enhancers in the TRAF1 locus and the ERAP2/LNPEP locus. Bar graphs summarize the results of 4 independent experiments. (A) Attenuation of the intergenic enhancer at the TRAF1 locus significantly reduced expression of PHF1, but not C5 (not shown). Off-target effects are monitored by showing that these gRNAs have no effect on HEB1 expression, but that gRNAs directed to an enhancer known to regulated HEB1 attenuates expression. (B) Attenuating the intergenic enhancer in the LNPEP/ERAP1 locus reduces expression of both LNPEP and ERAP2, but not the adjacent gene, CAST, nor the HEB1 gene (not shown). In each experiment, scrambled versions of the gRNAs had no effect on expression of the putative targets.
Objective: To develop a functional genomics pipeline to identify causal variants and their target genes on JIA risk haplotypes.
Design/Methods: We designed 180 bp oligonucleotides (“oligos”) representing 7,312 SNPs in LD with tag SNPs on JIA risk loci. Oligos were bar coded and cloned into a GFP-carrying plasmid vector behind a minimal promoter. We transfected oligo libraries into myeloid K562 cells to conduct a massively parallel reporter assay (MPRA) in which GFP expression for each SNP was compared to the common allele. We used 3D chromatin data to identify the chromatin loops harboring the enhancers carrying SNPs screened on MPRA. We used K562 cells stably transfected with the epigenome editing enzyme dCas-KRAB and gRNAs across the enhancer regions in the TRAF1 and LNPEP/ERAP2 loci, inducing dCas-KRAB with doxycycline and using qrtPCR to query expression levels of genes within the same chromatin loops.
Results: We identified n=44 SNPs that showed a significant difference in gene expression (FC > 2.0, FDR = 0.05) compared to the common allele. After stimulating K562 cells with IFNγ (250 ng/ml), we identified an additional 42 SNPs not identified in unstimulated cells. In many cases, we identified multiple alleles on the same haplotype, although these alleles were not always within the same functional element (e.g., intergenic vs intronic enhancers). Ablating the intergenic enhancer harboring rs10985080 on the TRAF1 haplotype using CRISPRi significantly reduced the expression of PHF1, but not C5, which is in the same chromatin loop. Using a similar approach for an intergenic enhancer in the LNPEP/ERAP2 locus, we found reduced expression of both LNPEP and ERAP2 but not CAST.Conclusion(s): We demonstrate a systematic method for identifying causal variants on JIA risk haplotypes based on observed functional properties. We also demonstrate that, once these variants are identified, target genes can be quickly identified using both publicly available 3D chromatin data and additional functional assays such as CRISPRi.
CRISPRi identifies target genes of enhancers harboring JIA-associated variants detected on MPRA
 For each experiment, 4 gRNAs were targeted to the functional regions of intergenic enhancers in the TRAF1 locus and the ERAP2/LNPEP locus. Bar graphs summarize the results of 4 independent experiments. (A) Attenuation of the intergenic enhancer at the TRAF1 locus significantly reduced expression of PHF1, but not C5 (not shown). Off-target effects are monitored by showing that these gRNAs have no effect on HEB1 expression, but that gRNAs directed to an enhancer known to regulated HEB1 attenuates expression. (B) Attenuating the intergenic enhancer in the LNPEP/ERAP1 locus reduces expression of both LNPEP and ERAP2, but not the adjacent gene, CAST, nor the HEB1 gene (not shown). In each experiment, scrambled versions of the gRNAs had no effect on expression of the putative targets.
For each experiment, 4 gRNAs were targeted to the functional regions of intergenic enhancers in the TRAF1 locus and the ERAP2/LNPEP locus. Bar graphs summarize the results of 4 independent experiments. (A) Attenuation of the intergenic enhancer at the TRAF1 locus significantly reduced expression of PHF1, but not C5 (not shown). Off-target effects are monitored by showing that these gRNAs have no effect on HEB1 expression, but that gRNAs directed to an enhancer known to regulated HEB1 attenuates expression. (B) Attenuating the intergenic enhancer in the LNPEP/ERAP1 locus reduces expression of both LNPEP and ERAP2, but not the adjacent gene, CAST, nor the HEB1 gene (not shown). In each experiment, scrambled versions of the gRNAs had no effect on expression of the putative targets.