Infectious Diseases
Category: Abstract Submission
Infectious Diseases: MIS-C & Kawasaki Disease
539 - Predicting IVIG Resistance in Kawasaki Disease with Machine Learning Algorithms augmented with Synthetic Sampling
Sunday, April 24, 2022
3:30 PM - 6:00 PM US MT
Poster Number: 539
Publication Number: 539.327
Publication Number: 539.327
Arthur Chang, John R. Oishei Children's Hospital, buffalo, NY, United States; Mark Hicar, Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo, Buffalo, NY, United States

Arthur Chang, MD
Fellow
John R. Oishei Children's Hospital
SUNY The State University of New York
buffalo, New York, United States
Presenting Author(s)
Background: Approximately 20% of children with Kawasaki Disease (KD), will be resistant to one dose of IVIG. Previous attempts to develop a score to predict which children are at risk largely do not work outside of Japan. Machine learning offers a novel way to predict IVIG resistance. Furthermore, augmenting limited data with synthetically derived data can improve performance. By training algorithms on an American KD dataset, a prediction algorithm and new insights can be gained.
Objective: Test various machine learning algorithms to attempt to predict which children, in an American dataset, are likely to be IVIG resistant. Interrogate the algorithm to identify insights as to what drives a models’ prediction on IVIG resistance.
Design/Methods: A dataset derived from previous work in western New York and publically available results from the pediatric heart network trial on pulse steroid therapy in Kawasaki Disease were used to train and test various algorithms available within scikit learn in a Jupyter notebook. Data was preprocessed for consistency, variables with significant (>30%) missing data were dropped, variables with < 30% missing data were imputed using the mean of the variable, categorical data was dummied, and continuous data were binned using kbins. We utilized two methods for handling class imbalance, class weighting and SMOTE. Algorithms were trained and best hyper-parameters were validated using nested cross validation. Results were evaluated on unseen folds of the data and using Shapley Additive Explanations we evaluated feature importances. Common performance statistics of algorithms were generated and learning curves were assessed to determine future model improvements. The experiment was repeated 4 other times with other randomization seeds.
Results: Synthetic weighting largely outperformed standard class weighting in most algorithms with respect to the receiver operator curve’s area under the curve (ROC-AUC). The random forest classifier had a mean sensitivity of 0.247, specificity of 0.900, and ROC-AUC of 0.686. The gradient boosting classifier had a mean sensitivity of 0.160, specificity of 0.894, and ROC-AUC of 0.618. Other algorithms tested, like the logistic regression classifier, support vector machine classifier, Naïve Bayes classifier, and Adaboost classifier did not perform as well. Learning curve analysis suggests, in some cases, more data will improve performance.Conclusion(s): These algorithms only performed as well as previous conventional scoring systems. However learning curve analysis suggests further refinement and more data will likely improve performance in American children.
Figure 1: Schematic representation of methods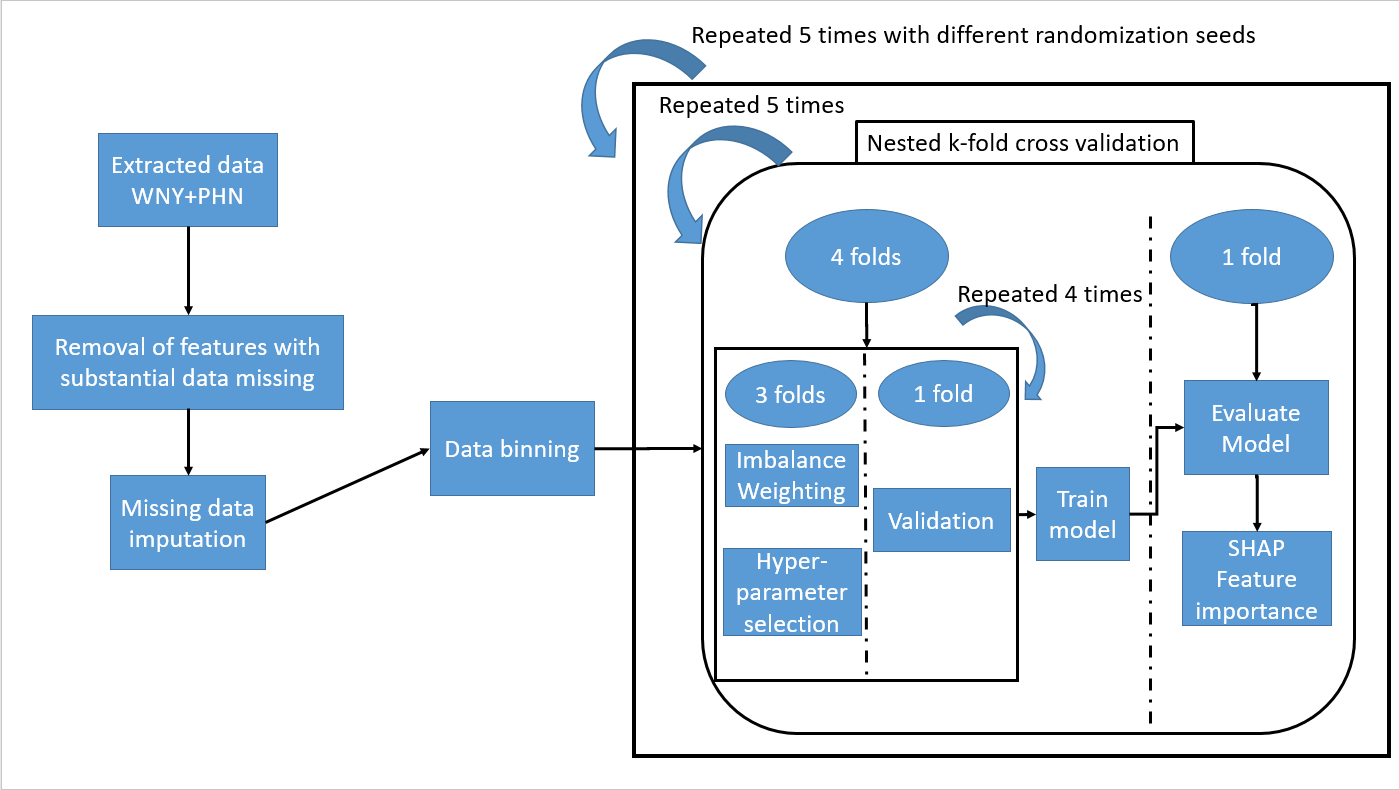 Data from our region (WNY) and extracted data from the Pediatric Heart Network study on Kawasaki Disease (PHN) were processed and binned before training with a nested k-fold cross validation strategy using both conventional class weighting and synthetic data weighting. The process was repeated a total of 5 times with different seeds to ensure results were replicable.
Data from our region (WNY) and extracted data from the Pediatric Heart Network study on Kawasaki Disease (PHN) were processed and binned before training with a nested k-fold cross validation strategy using both conventional class weighting and synthetic data weighting. The process was repeated a total of 5 times with different seeds to ensure results were replicable.
Table 1: parameter candidates tested in cross validation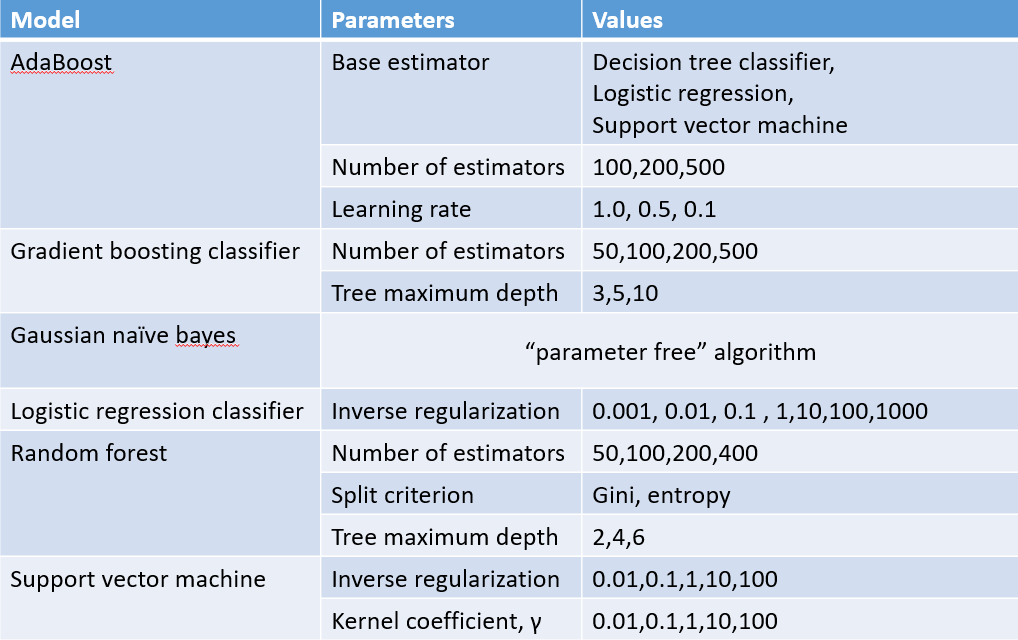
Objective: Test various machine learning algorithms to attempt to predict which children, in an American dataset, are likely to be IVIG resistant. Interrogate the algorithm to identify insights as to what drives a models’ prediction on IVIG resistance.
Design/Methods: A dataset derived from previous work in western New York and publically available results from the pediatric heart network trial on pulse steroid therapy in Kawasaki Disease were used to train and test various algorithms available within scikit learn in a Jupyter notebook. Data was preprocessed for consistency, variables with significant (>30%) missing data were dropped, variables with < 30% missing data were imputed using the mean of the variable, categorical data was dummied, and continuous data were binned using kbins. We utilized two methods for handling class imbalance, class weighting and SMOTE. Algorithms were trained and best hyper-parameters were validated using nested cross validation. Results were evaluated on unseen folds of the data and using Shapley Additive Explanations we evaluated feature importances. Common performance statistics of algorithms were generated and learning curves were assessed to determine future model improvements. The experiment was repeated 4 other times with other randomization seeds.
Results: Synthetic weighting largely outperformed standard class weighting in most algorithms with respect to the receiver operator curve’s area under the curve (ROC-AUC). The random forest classifier had a mean sensitivity of 0.247, specificity of 0.900, and ROC-AUC of 0.686. The gradient boosting classifier had a mean sensitivity of 0.160, specificity of 0.894, and ROC-AUC of 0.618. Other algorithms tested, like the logistic regression classifier, support vector machine classifier, Naïve Bayes classifier, and Adaboost classifier did not perform as well. Learning curve analysis suggests, in some cases, more data will improve performance.Conclusion(s): These algorithms only performed as well as previous conventional scoring systems. However learning curve analysis suggests further refinement and more data will likely improve performance in American children.
Figure 1: Schematic representation of methods
 Data from our region (WNY) and extracted data from the Pediatric Heart Network study on Kawasaki Disease (PHN) were processed and binned before training with a nested k-fold cross validation strategy using both conventional class weighting and synthetic data weighting. The process was repeated a total of 5 times with different seeds to ensure results were replicable.
Data from our region (WNY) and extracted data from the Pediatric Heart Network study on Kawasaki Disease (PHN) were processed and binned before training with a nested k-fold cross validation strategy using both conventional class weighting and synthetic data weighting. The process was repeated a total of 5 times with different seeds to ensure results were replicable.Table 1: parameter candidates tested in cross validation

