Neonatal GI Physiology & NEC
Category: Abstract Submission
Neonatal GI Physiology & NEC II
497 - Chronicling The Development Of The Neonatal Gut Microbiome: A Pilot Study
Monday, April 25, 2022
3:30 PM - 6:00 PM US MT
Poster Number: 497
Publication Number: 497.425
Publication Number: 497.425
Shubham M. Bakshi, Tufts Childrens Hospital, Revere, MA, United States; Jill Maron, Women & Infants Hospital of Rhode Island, Providence, RI, United States; Rachana Singh, Tufts Childrens Hospital, Boston, MA, United States; George Weinstock, Jackson Laboratory, Farmington, CT, United States; Thi Dong-binh Tran, The Jackson Laboratory For Genomic Medicine, Farmington, CT, United States

Shubham M. Bakshi, MD
Fellow
Tufts Childrens Hospital
Revere, Massachusetts, United States
Presenting Author(s)
Background: Neonatal gut microbial colonization plays a defining role in both the short- and long-term health of the developing infant. Biospecimens, such as saliva and stool, have the potential to provide an accurate view of ongoing neonatal gut colonization patterns through simultaneous genomic evaluations early in life.
Objective: To chronicle evolving salivary and stool microbial profiles in the first two weeks of life and explore associations between neonatal factors and early microbial colonization.
Design/Methods: In this novel, pilot, observational study, six stool and saliva samples were concurrently collected from each of the twenty subjects in first two weeks of life. 16s rRNA V1-V3 sequencing was performed and clustered to form an operational taxonomic unit (OTU). Centroid sequences for each OTU were mapped to correlate the taxonomic breakdown to the genus level. Alpha and beta diversity were analyzed based upon sex, mode of delivery, antibiotic exposure, presence of a central line, level of respiratory support and other morbidities.
Results: The first saliva and stool samples had the highest microbial diversity. Overall, saliva had higher alpha diversity in females as compared to males (p < 0.05) and in infants born via vaginal delivery as compared to infants born via cesarean delivery, independent of sex (p < 0.05). Subjects that had both maternal and neonatal antibiotic exposure had the highest alpha diversity (p < 0.05). Subjects who had no antibiotic exposure had lower overall alpha diversity in both stool and saliva as compared to subjects who had any antibiotic exposure (maternal or neonatal). Stool had higher alpha diversity in subjects with UVC as compared to PICC lines or no central lines (p < 0.05). Beta diversity between saliva and stool were dissimilar (Adonis R2= 0.062, and Anosim R= 0.1953, p < 0.05). Staphylococcus was the dominant genus in both stool and saliva, however, microbial composition of saliva and stool was dissimilar.Conclusion(s): Maternal and neonatal antibiotic exposure, sex, mode of delivery and presence of central line alter the neonatal microbiome. Salivary microbial profiling does not appear to be predictive of stool microbial profiles in the first two weeks of life. Associations with morbidities must be further explored.
Dr. Shubham Bakshi CVDr. Shubham Bakshi Resume.pdf
Figure 1: Alpha diversity over time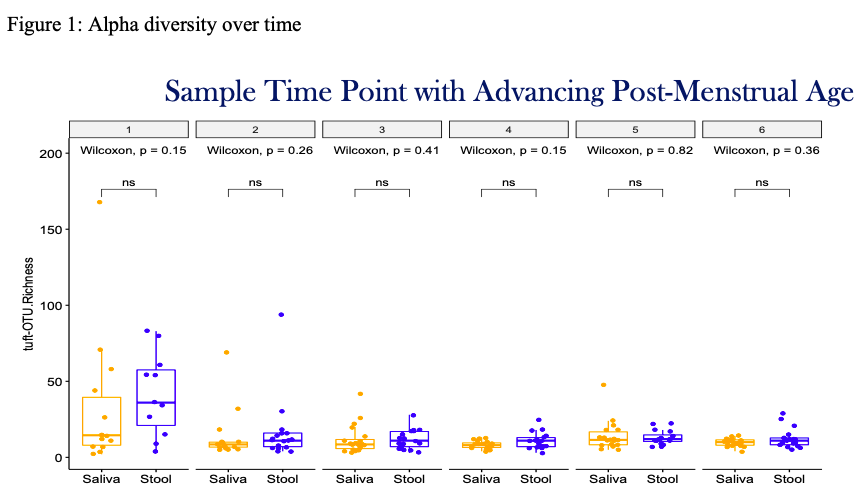
Objective: To chronicle evolving salivary and stool microbial profiles in the first two weeks of life and explore associations between neonatal factors and early microbial colonization.
Design/Methods: In this novel, pilot, observational study, six stool and saliva samples were concurrently collected from each of the twenty subjects in first two weeks of life. 16s rRNA V1-V3 sequencing was performed and clustered to form an operational taxonomic unit (OTU). Centroid sequences for each OTU were mapped to correlate the taxonomic breakdown to the genus level. Alpha and beta diversity were analyzed based upon sex, mode of delivery, antibiotic exposure, presence of a central line, level of respiratory support and other morbidities.
Results: The first saliva and stool samples had the highest microbial diversity. Overall, saliva had higher alpha diversity in females as compared to males (p < 0.05) and in infants born via vaginal delivery as compared to infants born via cesarean delivery, independent of sex (p < 0.05). Subjects that had both maternal and neonatal antibiotic exposure had the highest alpha diversity (p < 0.05). Subjects who had no antibiotic exposure had lower overall alpha diversity in both stool and saliva as compared to subjects who had any antibiotic exposure (maternal or neonatal). Stool had higher alpha diversity in subjects with UVC as compared to PICC lines or no central lines (p < 0.05). Beta diversity between saliva and stool were dissimilar (Adonis R2= 0.062, and Anosim R= 0.1953, p < 0.05). Staphylococcus was the dominant genus in both stool and saliva, however, microbial composition of saliva and stool was dissimilar.Conclusion(s): Maternal and neonatal antibiotic exposure, sex, mode of delivery and presence of central line alter the neonatal microbiome. Salivary microbial profiling does not appear to be predictive of stool microbial profiles in the first two weeks of life. Associations with morbidities must be further explored.
Dr. Shubham Bakshi CVDr. Shubham Bakshi Resume.pdf
Figure 1: Alpha diversity over time

