Critical Care
Category: Abstract Submission
3: Critical Care I
22 - Clinic Utility of Nasal Staphylococcus Aureus Screening by Polymerase Chain Reaction (PCR) in Pediatric Pneumonia
Friday, April 22, 2022
6:15 PM - 8:45 PM US MT
Poster Number: 22
Publication Number: 22.102
Publication Number: 22.102
Ana A. Delerme, Nationwide Children's Hospital, Columbus, OH, United States; Joshua Watson, Nationwide Children's Hospital and The Ohio State University, Columbus, OH, United States; Todd Karsies, Nationwide Children's Hospital, Columbus, OH, United States; Corey Toocheck, Premier Medical Associates, Monroeville, PA, United States

Ana A. Delerme, MD
Pediatric Hospitalist
Nationwide Children's Hospital
Columbus, Ohio, United States
Presenting Author(s)
Background: Pneumonia is a major cause of admission to the pediatric intensive care unit (PICU). Children who require respiratory support often receive empiric, broad-spectrum antibiotics including vancomycin to target Staphylococcus aureus (SA) while awaiting respiratory culture results. Studies in adult patients using nasal SA PCR swab demonstrate a high negative predictive value, suggesting colonization evaluation could be an effective tool for rapid de-escalation of empiric therapy. No studies regarding this association are available in the pediatric literature to date.
Objective: To determine the utility of SA PCR from nasal swab to predict lower respiratory tract (LRT) culture or pleural fluid culture in patients with pneumonia who require mechanical ventilation.
Design/Methods: We prospectively enrolled patients 17 years old and younger with clinically diagnosed pneumonia and respiratory failure requiring mechanical ventilator support in the Nationwide Children’s Hospital PICU. Patients included had a LRT culture obtained via tracheal aspirate or bronchoalveolar lavage or had pleural fluid culture obtained and received empiric vancomycin within 24 hours of PICU admission. Nasal swabs were obtained for SA PCR using the Cepheid Xpert SA Complete assay, which detects the spa gene (indicating SA colonization) and the mecA and SCCmec genes (indicating methicillin resistance). Results of LRT or pleural fluid cultures served as the gold standard. We determined characteristics of the assay, including sensitivity, specificity, and positive and negative predictive values (PPV, NPV).
Results: We enrolled 81 patients, ages 12 days to 17 years old. LRT or pleural fluid cultures yielded SA in 20 (24.7%) patients: 15 were methicillin-susceptible SA (MSSA) and 5 were methicillin-resistant SA (MRSA). Nasal SA PCR detected SA colonization in 36 (44.4%) patients, including 19 of the 20 patients with positive SA culture (MSSA or MRSA). For predicting isolation SA in culture, the nasal SA PCR showed sensitivity 95.0%, specificity 72.1%, PPV 52.78% and NPV 97.8%. Regarding MRSA specifically, nasal SA PCR detected MRSA colonization in 9 (11.1%) patients, including 5 patients whose culture yielded MRSA. Thus, for predicting isolation of MRSA in culture, the nasal SA PCR showed sensitivity 100%, specificity 94.7%, PPV 55.6% and NPV 100%.Conclusion(s): Our data suggest that nasal SA PCR may be an effective tool to rule out SA and MRSA LRT infection in pediatric patients. This will allow rapid de-escalation of broad-spectrum antibiotics, thus avoiding unnecessary vancomycin exposure and its associated adverse effects.
Staphylococcus aureus (SA) results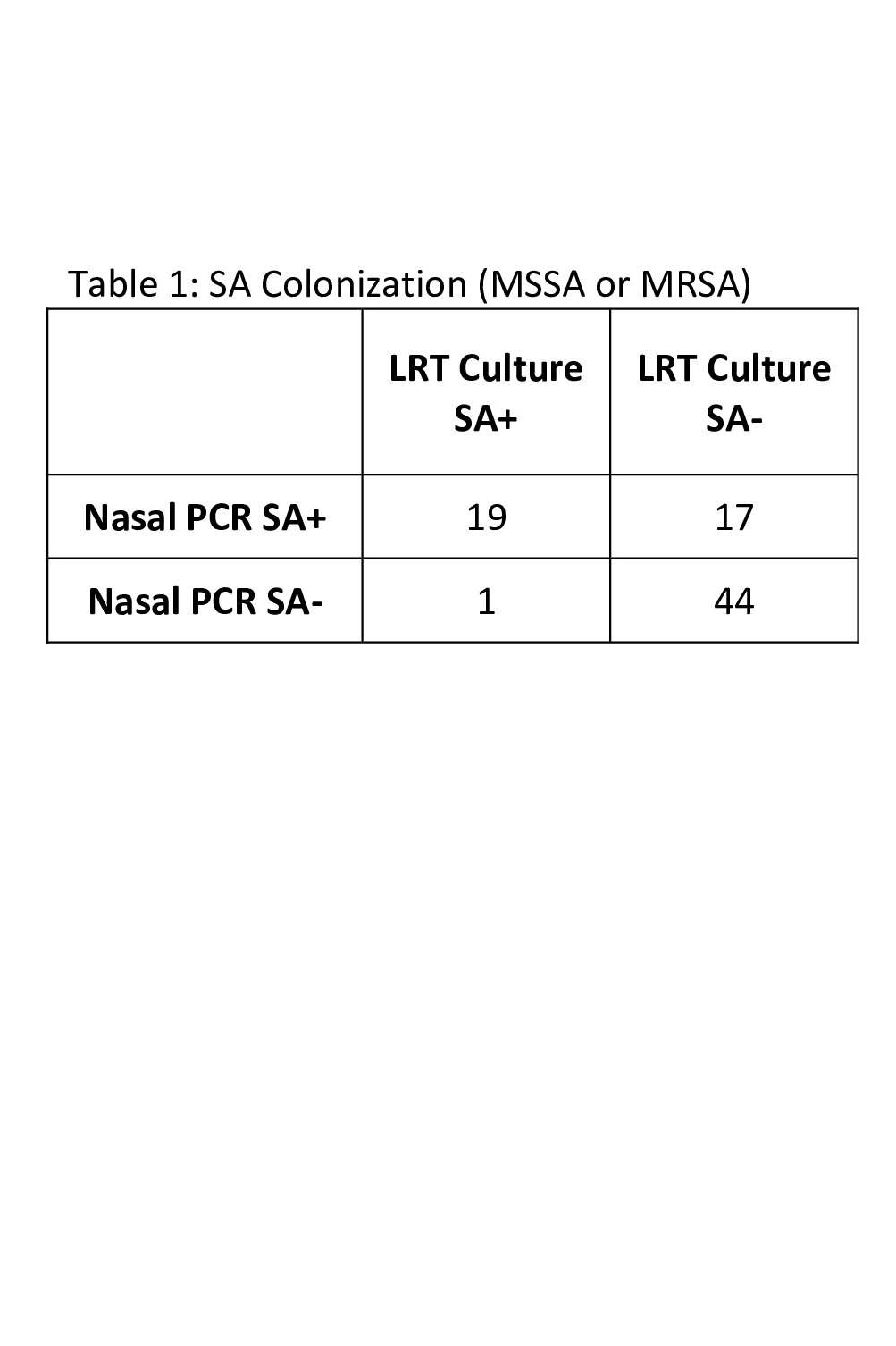 This table shows the number of patients in which SA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew any SA.
This table shows the number of patients in which SA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew any SA.
Methicillin Resistant Staphylococcus aureus (MRSA) Results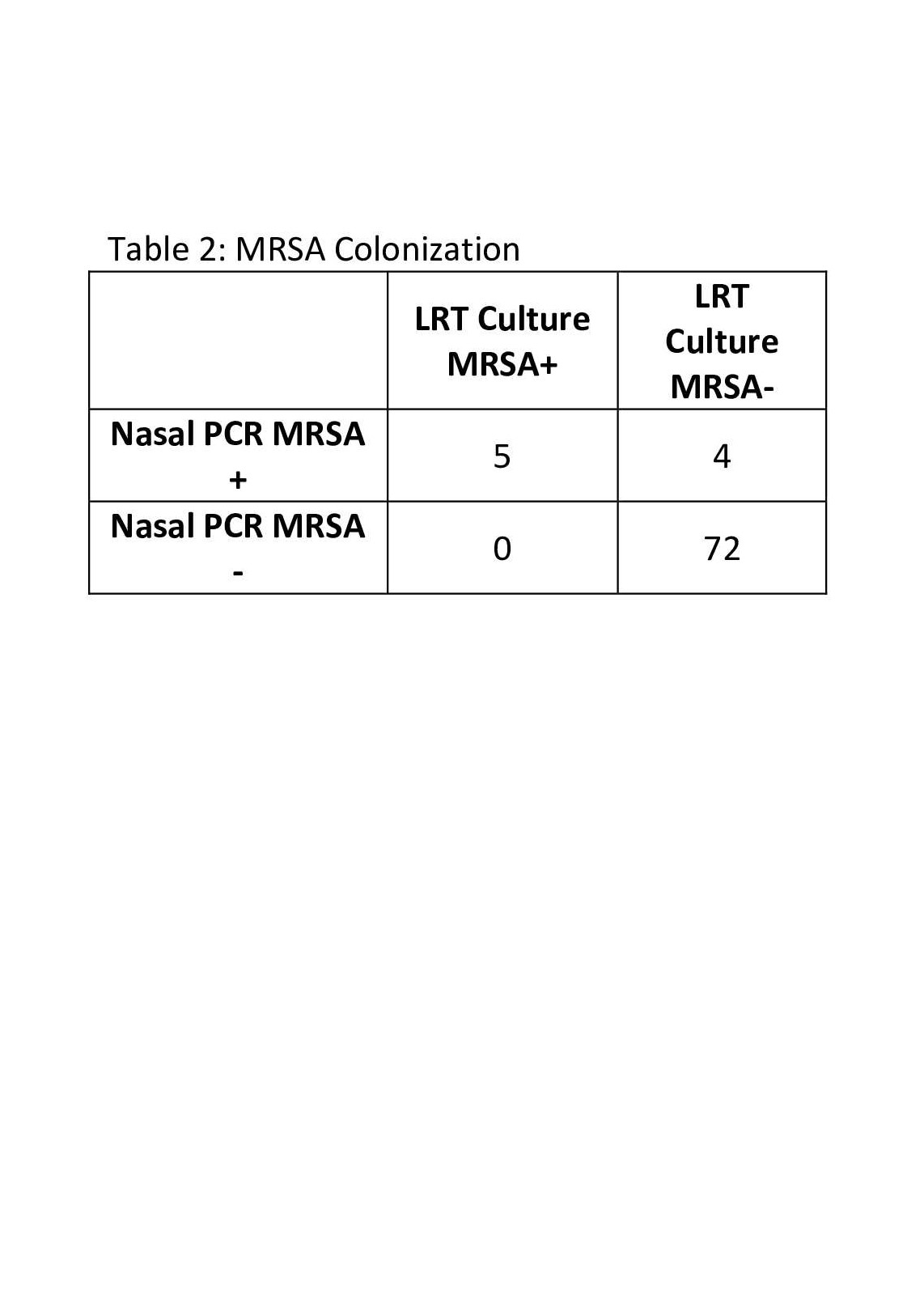 This table shows the number of patients in which MRSA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew MRSA specifically.
This table shows the number of patients in which MRSA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew MRSA specifically.
Objective: To determine the utility of SA PCR from nasal swab to predict lower respiratory tract (LRT) culture or pleural fluid culture in patients with pneumonia who require mechanical ventilation.
Design/Methods: We prospectively enrolled patients 17 years old and younger with clinically diagnosed pneumonia and respiratory failure requiring mechanical ventilator support in the Nationwide Children’s Hospital PICU. Patients included had a LRT culture obtained via tracheal aspirate or bronchoalveolar lavage or had pleural fluid culture obtained and received empiric vancomycin within 24 hours of PICU admission. Nasal swabs were obtained for SA PCR using the Cepheid Xpert SA Complete assay, which detects the spa gene (indicating SA colonization) and the mecA and SCCmec genes (indicating methicillin resistance). Results of LRT or pleural fluid cultures served as the gold standard. We determined characteristics of the assay, including sensitivity, specificity, and positive and negative predictive values (PPV, NPV).
Results: We enrolled 81 patients, ages 12 days to 17 years old. LRT or pleural fluid cultures yielded SA in 20 (24.7%) patients: 15 were methicillin-susceptible SA (MSSA) and 5 were methicillin-resistant SA (MRSA). Nasal SA PCR detected SA colonization in 36 (44.4%) patients, including 19 of the 20 patients with positive SA culture (MSSA or MRSA). For predicting isolation SA in culture, the nasal SA PCR showed sensitivity 95.0%, specificity 72.1%, PPV 52.78% and NPV 97.8%. Regarding MRSA specifically, nasal SA PCR detected MRSA colonization in 9 (11.1%) patients, including 5 patients whose culture yielded MRSA. Thus, for predicting isolation of MRSA in culture, the nasal SA PCR showed sensitivity 100%, specificity 94.7%, PPV 55.6% and NPV 100%.Conclusion(s): Our data suggest that nasal SA PCR may be an effective tool to rule out SA and MRSA LRT infection in pediatric patients. This will allow rapid de-escalation of broad-spectrum antibiotics, thus avoiding unnecessary vancomycin exposure and its associated adverse effects.
Staphylococcus aureus (SA) results
 This table shows the number of patients in which SA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew any SA.
This table shows the number of patients in which SA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew any SA.Methicillin Resistant Staphylococcus aureus (MRSA) Results
 This table shows the number of patients in which MRSA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew MRSA specifically.
This table shows the number of patients in which MRSA colonization was detected by nasal PCR and those whose lower respiratory tract (LRT) culture grew MRSA specifically.