Genomics/Epigenomics
Category: Abstract Submission
2: Genomics/Epigenomics II
11 - A Machine Learning Classifier to Prioritize Exome Reanalysis and Increase Rates of Rare Disease Diagnosis
Friday, April 22, 2022
6:15 PM - 8:45 PM US MT
Poster Number: 11
Publication Number: 11.110
Publication Number: 11.110
Austin A. Antoniou, Nationwide Children's Hospital, Columbus, OH, United States; Robert Schuetz, Nationwide Children's Hospital, Columubs, OH, United States; Bimal P. Chaudhari, Nationwide Children's Hospital, Columbus, OH, United States

Austin A. Antoniou, PhD (he/him/his)
Postdoc
Nationwide Children's Hospital
Columbus, Ohio, United States
Presenting Author(s)
Background: Over 25 million Americans are affected by rare diseases most of which have a genetic component. Exome Sequencing (ES) and Genome Sequencing (GS), among the most comprehensive tests which can be used to diagnose rare genetic disorders, have a diagnostic rate of 25-40%. However, as knowledge linking the genome to diseases grows, and as individual patients’ phenotypes evolve, previously unsolved cases may become solvable.
Objective: The goal of this work is to build a machine-learning classifier which leverages case-level and demographic data in tandem with disease ranking scores to classify and prioritize the cases which are most likely to benefit from focused human reanalysis effort.
Design/Methods: We considered 611 patients who received clinical ES and consented for further research. They were divided into three groups: solved (195), potentially solvable (337), and unlikely to be solvable (79). Setting aside the potentially solvable population, we built a classifier to distinguish the solved cases (positive class) from the unlikely solvable cases (negative class). Patient age, sex, and time since most recent analysis were used as case-level features to inform the model. Additional features included the top 25 scores from a modified version of LIRICAL, a likelihood ratio-based tool for ranking variants given a patient’s Variant Call Format file and a list of relevant Human Phenotype Ontology terms. An 80:20 train-test split was made, and 41 solved and 26 unlikely solvable research GS cases were added to the training data. Logistic regression, random forest, K nearest neighbors, support vector, and gradient boosting classifiers, with optional SMOTE oversampling and principal component decomposition, were selected by a parameter grid search and 5-fold cross validation (Figure 1). The models of each type with the best F1 score were used to build a voting classifier.
Results: The grid search yielded a voting classifier with 89.5% recall and 75.6% precision on the test set. The model called 259 of the 337 potentially solvable cases positive, meaning they are likely solvable. Adjusting for the model’s precision, 195 of these cases should be solvable. A physician determined via manual chart review that at least 4 of the 10 top-scoring cases identified by the model are viable candidates for formal reanalysis on a clinical basis.Conclusion(s): A model informed by both variant and case-level data can aid in better allocating human effort toward genetic test reanalysis. Including features such as the time since previous analysis allows the opportunity for virtually all cases to eventually be re-evaluated.
Voting Reanalysis Classifier Flowchart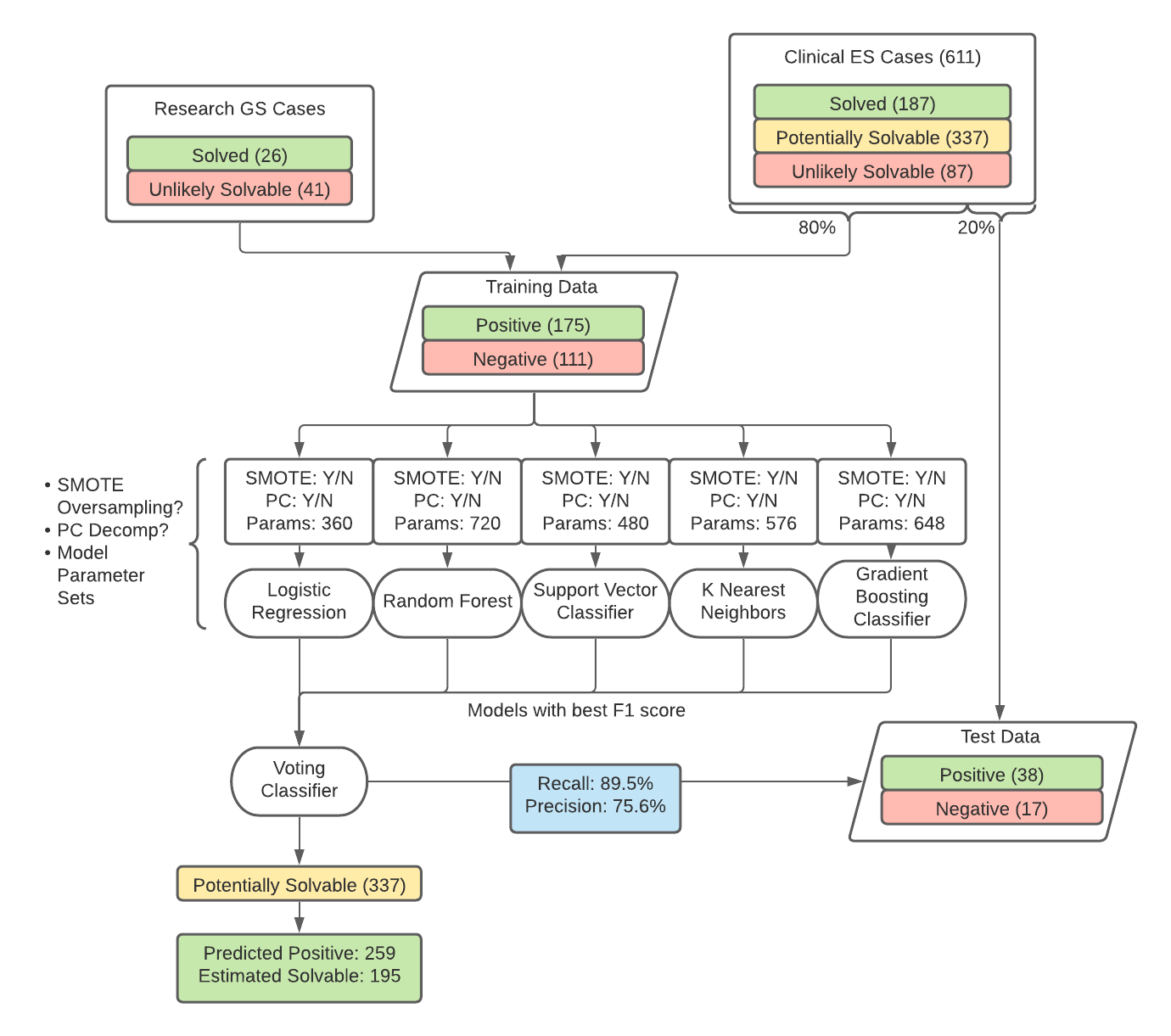 A visual summary of the process used to build the reanalysis classifier and predict which potentially solvable cases are most likely to be solvable.
A visual summary of the process used to build the reanalysis classifier and predict which potentially solvable cases are most likely to be solvable.
Objective: The goal of this work is to build a machine-learning classifier which leverages case-level and demographic data in tandem with disease ranking scores to classify and prioritize the cases which are most likely to benefit from focused human reanalysis effort.
Design/Methods: We considered 611 patients who received clinical ES and consented for further research. They were divided into three groups: solved (195), potentially solvable (337), and unlikely to be solvable (79). Setting aside the potentially solvable population, we built a classifier to distinguish the solved cases (positive class) from the unlikely solvable cases (negative class). Patient age, sex, and time since most recent analysis were used as case-level features to inform the model. Additional features included the top 25 scores from a modified version of LIRICAL, a likelihood ratio-based tool for ranking variants given a patient’s Variant Call Format file and a list of relevant Human Phenotype Ontology terms. An 80:20 train-test split was made, and 41 solved and 26 unlikely solvable research GS cases were added to the training data. Logistic regression, random forest, K nearest neighbors, support vector, and gradient boosting classifiers, with optional SMOTE oversampling and principal component decomposition, were selected by a parameter grid search and 5-fold cross validation (Figure 1). The models of each type with the best F1 score were used to build a voting classifier.
Results: The grid search yielded a voting classifier with 89.5% recall and 75.6% precision on the test set. The model called 259 of the 337 potentially solvable cases positive, meaning they are likely solvable. Adjusting for the model’s precision, 195 of these cases should be solvable. A physician determined via manual chart review that at least 4 of the 10 top-scoring cases identified by the model are viable candidates for formal reanalysis on a clinical basis.Conclusion(s): A model informed by both variant and case-level data can aid in better allocating human effort toward genetic test reanalysis. Including features such as the time since previous analysis allows the opportunity for virtually all cases to eventually be re-evaluated.
Voting Reanalysis Classifier Flowchart
 A visual summary of the process used to build the reanalysis classifier and predict which potentially solvable cases are most likely to be solvable.
A visual summary of the process used to build the reanalysis classifier and predict which potentially solvable cases are most likely to be solvable.